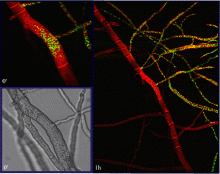
In nature, a cell will encounter many potential fusion partners with variable degrees of genetic similarity. Therefore, as an attempt to judge between genetic differences that can be either beneficial or detrimental for organismal fitness, Neurospora crassa cells have developed self/non-self recognition mechanisms that are put in place upon the imminence of cell-cell communication and fusion. Our group has shown that among a wild population of Neurospora isolates, a long-distance kind recognition system that functions at the level of germling communication determines the successfulness of interaction between two germlings. This allorecognition system involves a highly divergent locus on chromosome 5, which was appropriately dubbed as ‘determinant of communication’ (doc) and encompasses genes doc-1, doc-2 and doc-3. Five communication group haplotypes (CGH) were identified in the analyzed wild population and only cells with a similar CGH are able to chemotropically interact with each other.

Communication groups (CGs) within a Neurospora crassa population from Louisiana. (Heller et al, 2016)
In mature hyphae, nonself recognition during heterokaryon formation in filamentous fungi is regulated by genetic loci, termed het (for heterokaryon incompatibility) loci. Three het loci, mat, het-c, and het-6, have been cloned and characterized at the molecular level and have been shown to encode genes with highly polymorphic allelic specificity domains.
Molecular mechanism of nonself recognition
At the het-c locus, nonself recognition is dependent on the het-c haplotype. Nonself recognition requires both allelic (het-c) and non-allelic interactions (between het-c and pin-c). Individuals carrying het-c1 pin-c1 specificity are incompatible with individuals carrying het-c2 pin-c2 or het-c3 pin-c3 specificity. The specificity domain of het-c and pin-c are highly polymorphic (i.e. pin-c alleles are ~50% identical and DNA and protein level). It is of interest to note that strains of alternative het-c haplotype are completely inter-fertile, indicating that heterokaryon incompatibility is suppressed during sexual development. We are currently assessing protein-protein interactions between HET-C and PIN- C and determining the protein regions required for this interaction.
Molecular Mechanism of Signal Transduction & Death
A number of mutants have been identified that affect nonself recognition and PCD in Neurospora. These include a putative transcription factor, vib-1. Our current research efforts are aimed at understanding how nonself recognition triggers programmed cell death via activation of VIB-1. We are currently using microarray technology and chromatin immunoprecipitation to identify targets of VIB-1 to gain an understanding of how the hyphal compartments are compartmentalized and killed.
Evolution of Nonself Recognition
In vertebrate species, self/non-self recognition relies on the major histocompatibility complex (MHC), which is an array of genetic loci that generate proteins important in pathogen recognition and activation of defense mechanisms.Alleles at MHC loci show long term persistence, such that an allele from one species is often more closely related to an allele in a different species, a pattern termed trans-species polymorphisms. Self/non-self recognition during sexual reproduction in many plant species is mediated by the gametophytic or sporophytic self-incompatibility locus, S, which elicits recognition and rejection of self-pollen. Alleles at the S locus are also extremely polymorphic and show trans-species polymorphisms. Allelic polymorphisms at some het loci in Neurospora show evidence of balancing selection, i.e. alleles are highly polymorphic, alleles of alternative specificity are equally frequent in populations and show trans-species polymorphisms. We are currently taking a genomic approach to identify all of the nonself recognition loci in the genome of N. crassa and to compare evolutionary trajectories of these loci in filamentous fungi.
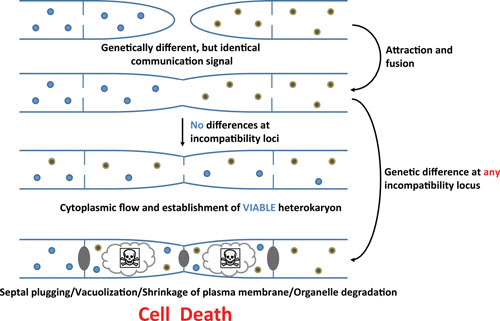
Heterokaryosis and its possible outcomes. (Daskalov et al, 2016)